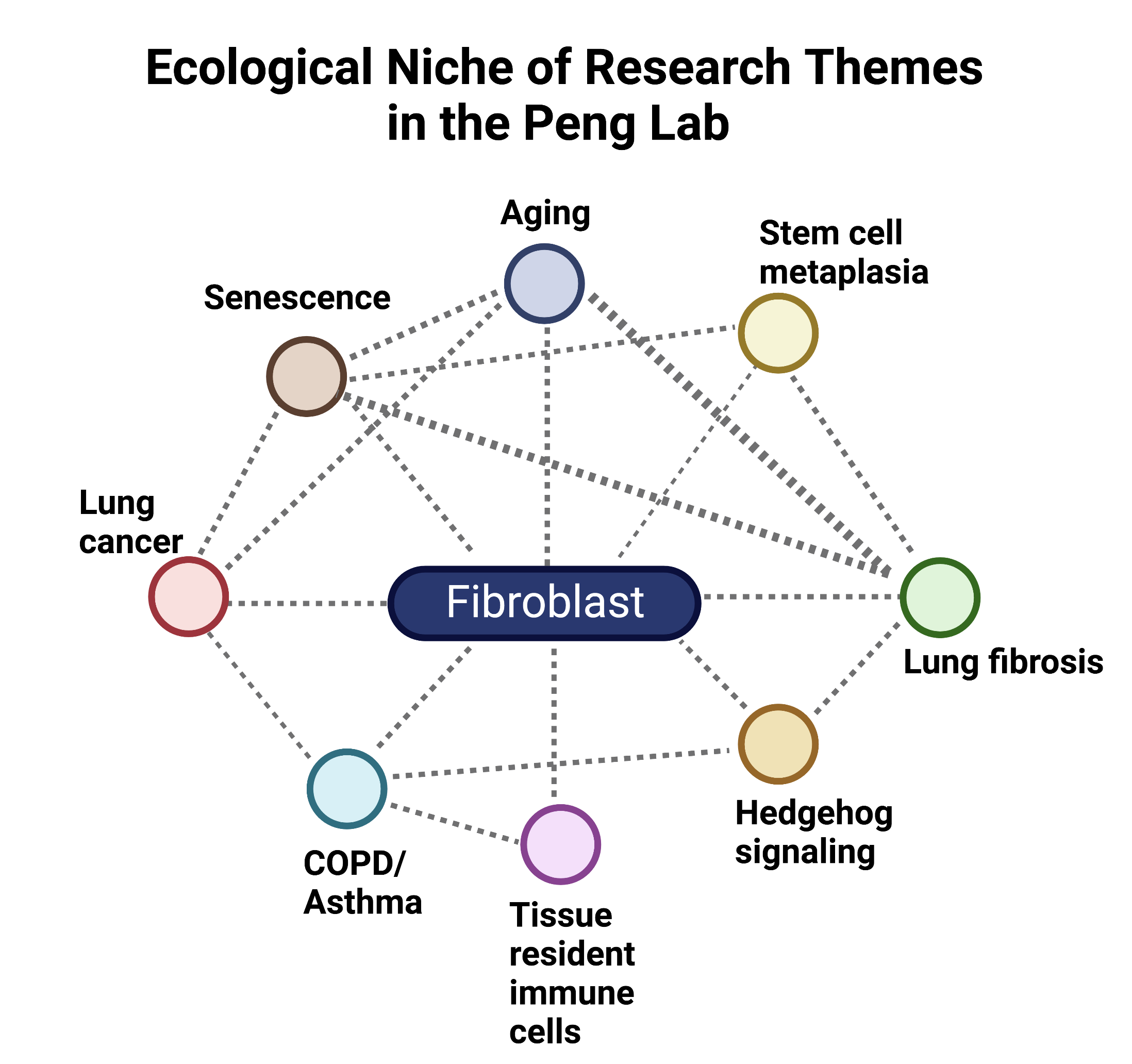
The Peng Lab conceptualizes the tissue as an ecological niche with fibroblasts at the center. Along those lines, our research themes center around the fibroblast as the key node in driving diverse cellular processes and diseases that are interrelated. The diagram below represents the evolving relationships we are exploring driven by fibroblasts. We are always looking for new circles/ideas to add to this diagram.
micro-CT scan of a healthy and emphysematous lung where fibroblast signaling is altered.
Deciphering the role of fibroblast in driving disease phenotypes
Our lab is interested in the development of new murine genetic models to characterize the role of fibroblasts in driving disease phenotypes. By utilizing genetic tools where we can modify fibroblasts in pre-clinical disease models, we can dissect how fibroblasts drive the pathologic features that define diseases such as COPD/emphysema, lung fibrosis, cancer, asthma, and etc.
Single cell RNA sequencing of the human lung
Stromal aging model
Developing single-cell pipelines to dissect Stromal heterogeneity
We are focused on understanding how certain developmental pathways subdivide fibroblast subpopulations based on location and function. We are performing single-cell RNA sequencing using both the 10X Genomics and C1 Fluidigm platform to better understand how fibroblast transcriptomes segregate based on location and cellular behavior in mouse and human.
Establishment of organoid culture system to study tissue-tissue interaction
Our lab is interested in dissecting paracrine interactions that maintain normal tissue homeostasis, and modeling these behaviors in vitro. We have established organoid culture systems to study how various fibroblast subsets support epithelial stem/progenitors with both mouse and human derived samples. The goal is to identify fibroblast-derived factors that regulate epithelial stem/progenitor behavior and vice versa.